Secrets from beyond extinction: the Tasmanian tiger
The entire thylacine genome has now been sequenced, revealing the apex marsupial predator was in poor genetic health and may have struggled to fight disease had it survived.
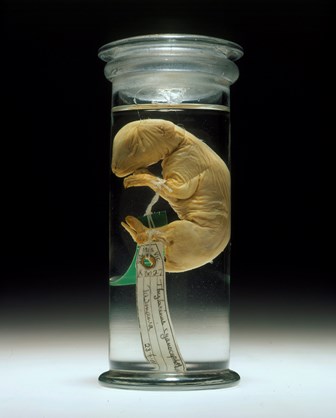
Floating in a small jar of alcohol sits one of Australia's rarest specimens.
The jar, labelled collection number C5757, holds a juvenile Tasmanian tiger or thylacine, one of the best-preserved extinct species, now held in Museums Victoria's Collection.
As the animal became rarer, museums everywhere clambered to have a thylacine on show, and they are now its last refuge after being hunted to extinction in 1936.
Using techniques never imagined when the last thylacine died in Hobart Zoo last centry, a team led by the University of Melbourne have now sequenced the genome of the Tasmanian tiger (Thylacinus cynocephalus), making it one of the most complete genetic blueprints for an extinct animal.
For project leader Professor Andrew Pask, the thylacine is his labour of love. Over ten years ago, he and an international team first resurrected a Tasmanian tiger gene from preserved pelt, but the DNA was too fragmented to obtain the whole genome.
So, they searched museums' world-wide databases and found specimen C5757 in Museums Victoria’s collection – a young thylacine pup. Because the Tasmanian tiger was a marsupial, which are mammals with a pouch, this pup specimen could be preserved in its entirety, allowing the research team to extract DNA and use cutting-edge techniques to sequence the thylacine genome.
Associate Professor Andrew Pask says the results provide the first full genetic blueprint of the largest Australian apex predator to survive into the modern era.
"The genome allows us to confirm the thylacine's place in the evolutionary tree. The Tasmanian tiger belongs in a sister lineage to the Dasyuridae, the family which includes the Tasmanian Devil and the dunnart," says Associate Professor Pask, from the School of Biosciences.
Importantly, the genome has also revealed the poor genetic health, or low genetic diversity, the thylacine experienced before it was over-hunted. The Tasmanian Devil is now also facing a 'genetic bottleneck' which is a likely result of their genetic isolation from mainland Australia for the last 10,000 to 13,000 years.
However, the genome analysis suggests that both animals were experiencing low genetic diversity before they became isolated on Tasmania. This, in turn, suggests that Tasmanian tigers may have faced similar environmental problems to the Devils, had they survived, such as a difficulty overcoming disease.
"Our hope is that there is a lot the thylacine can tell us about the genetic basis of extinction to help other species," Associate Professor Pask says.
"As this genome is one of the most complete for an extinct species, it is technically the first step to 'bringing the thylacine back', but we are still a long way off that possibility.
"We would still need to develop a marsupial animal model to host the thylacine genome, like work conducted to include mammoth genes in the modern elephant. But knowing the Tasmanian tiger was facing limited genetic diversity before extinction means it would still have struggled similarly to the Tasmanian Devil if it had survived."
The genome provides other important new insights into the biology of this truly unique marsupial.
The thylacine is often described as a long dog with stripes, because it had a long, stiff tail and a big head. A fully grown thylacine could measure 180cm from the tip of the nose to the tip of the tail and stand 58cm high.
Its thick black stripes extended from the shoulders to the base of the tail.
Like the dingo, the thylacine was a very quiet animal. But they were reported to be relentless hunters who pursed their prey until it was exhausted.
Scientists consider the thylacine and the dingo as one of the best examples of 'convergent evolution', the process whereby organisms that are not closely related independently evolve to look the same as a result of having to adapt to similar environments or ecological niches.
It appears that because of their hunting technique and diet of fresh meat, their skulls and body shape became extremely similar.
Working with Dr Christy Hipsley from Museums Victoria, the team analysed the characteristics of the thylacine’s skull - such as eye, jaw and snout shape.
"We found the Tasmanian tiger had a more similar skull shape to the red fox and gray wolf than to its closest relatives," Dr Hipsley says.
"The fact these groups have not shared a common ancestor since the Jurassic period makes this an astounding example of convergence between distantly related species."
Associate Professor Pask says that the thylacine looked almost like a dingo with a pouch.
"When we looked at the basis for this convergent evolution, we found it wasn't actually the genes that produced the same skull and body shape, but the control regions around them that turn genes 'on and off' at different stages of growth.
"This reveals a whole new understanding of the process of evolution. We can now explore these regions of the genome to help understand how two species converge on the same appearance, and how the process of evolution works."
In this case, it seems the need to hunt led the thylacine to transform its appearance into one similar to the wolf over the past 160 million years.
Scientists can now start to understand the genetics that have driven this process and uncover more about the biology of this unique marsupial apex predator.
The research team also included scientists from the University of Munster, Museums Victoria, University of Adelaide and University of Connecticut. Some of the work was funded by the Research @ Melbourne Accelerator Program.
This article was written by Dr Nerissa Hannink, University of Melbourne.
This article was first published on Pursuit. Read the original article.
Join Dr Christy Hipsley as she describes the 3D digital imaging techniques used to reconstruct the thylacine’s unique biology at Melbourne Museum on the Tuesday 20 March 2018.